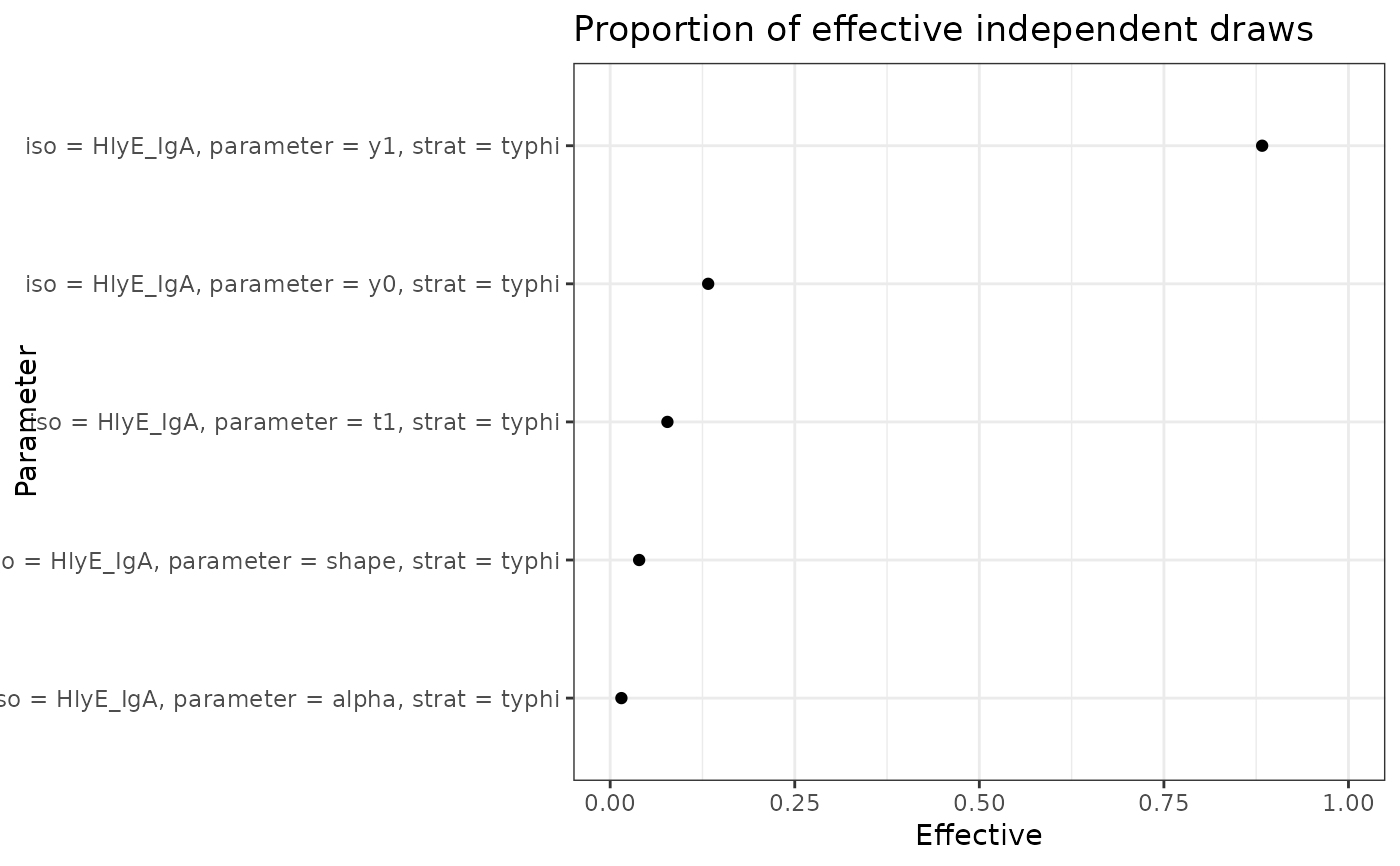
plot_jags_effect() takes a list output from run_mod()
to create summary diagnostics for each chain run in the mcmc estimation.
Defaults will produce every combination of antigen/antibody, parameters,
and stratifications, unless otherwise specified. At least 2 chains are
required to run function.
Antigen/antibody combinations and stratifications will vary by analysis.
The antibody dynamic curve includes the following parameters:
y0 = baseline antibody concentration
y1 = peak antibody concentration
t1 = time to peak
r = shape parameter
alpha = decay rate
Arguments
- data
A list outputted from run_mod().
- iso
Specify character string to produce plots of only a specific antigen/antibody combination, entered with quotes. Default outputs all antigen/antibody combinations.
- param
Specify character string to produce plots of only a specific parameter, entered with quotes. Options include:
y0= posterior estimate of baseline antibody concentrationy1= posterior estimate of peak antibody concentrationt1= posterior estimate of time to peakr= posterior estimate of shape parameteralpha= posterior estimate of decay rate
- strat
Specify character string to produce plots of specific stratification entered in quotes.
Value
A list of ggplot2::ggplot objects showing the proportion of effective samples taken/total samples taken for all parameter iso combinations. The estimate with the highest proportion of effective samples taken will be listed first.
Examples
data <- serodynamics::nepal_sees_jags_output
plot_jags_effect(data = data,
iso = "HlyE_IgA",
strat = "typhi")