Serocalculator package installation manual
UC Davis Seroepidemiology Research Group (SERG)
2023-11-21
Source:vignettes/installation.Rmd
installation.Rmd1. Introduction
Package serocalculator is written in programming language R and end user must have access to a working installation of R engine. This document describes the most common setup with R installed locally on the user’s computer. The screenshots refer to the classical R interface, but the package can also be opened in other Graphical User Interfaces for R like e.g. RStudio.
2. Installation steps
R is a free software program and can be downloaded from http://cran.r-project.org/. After downloading the appropriate version for your computer’s operating system, install R on your computer following the standard procedure applicable to the operating system. For Windows the file to be downloaded is the so-called base distribution: http://cran.r-project.org/bin/windows/base/.
2.1. Installing R
Start the R installer and follow the presented steps:
It is advised to have R installed in folder that does not contain spaces, therefore please adjust the destination location accordingly:
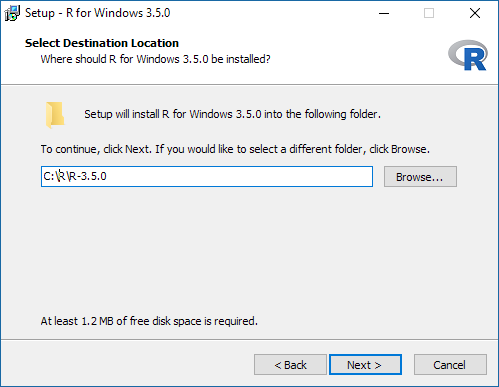
The serocalculator package is compatible with both the 32-bit and the 64-bit version of R. Choose the preferred platform (or both). If unsure install the 32-bit version only, however on compatible platforms the 64-bit version may provide better performance:
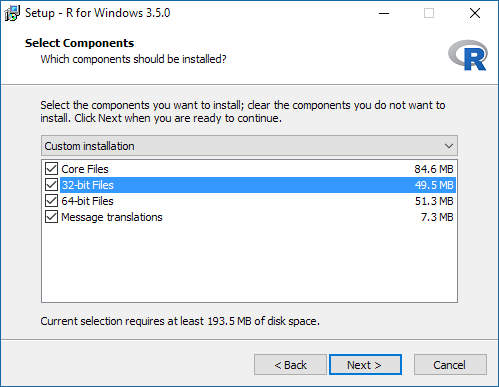
It is advised to select Registry entries in the next step for best experience:
R interpreter, when installed on Windows, can be invoked from the start menu folder named R.
Start the preferred version of R (if both the 32-bit: R i386 and the 64-bit: x64 are installed). Graphical user interface for R interpreter will start in a new window:
2.2. Installing serocalculator package
Since this is a new installation of R, the serocalculator package must be installed before first use. As of 09/20/2023, serocalculator is still in development. To install the development version, you must install the devtools package and then download serocalculator from GitHub.
install.packages("devtools")
devtools::install_github("ucd-serg/serocalculator")3. Post-installation
Successful installation can be confirmed by loading the package into the workspace and exploring help files and manuals distributed with the package:
# Load package "seroincidence".
library(serocalculator)
# Show R help for the package.
?serocalculator
# Show tutorial for the package.
vignette(topic = "tutorial", package = "serocalculator")Additionally, most package details can be found when executing the following commands:
# Show description.
packageDescription("serocalculator")## Package: serocalculator
## Type: Package
## Title: Estimating Infection Rates from Serological Data
## Version: 0.1.0.9000
## Date: 2022-03-29
## Authors@R: c( person(given = "Peter", family = "Teunis", email =
## "p.teunis@emory.edu", role = c("aut", "cph"), comment = "Author
## of the method and original code."), person(given = "Kristina",
## family = "Lai", role = c("aut")), person(given = "Kristen",
## family = "Aiemjoy", email = "kaiemjoy@ucdavis.edu", role =
## c("aut")), person(given = "Douglas Ezra", family = "Morrison",
## email = "demorrison@ucdavis.edu", role = c("aut", "cre")))
## Description: Translates antibody levels measured in cross-sectional
## population samples into estimates of the frequency with which
## seroconversions (infections) occur in the sampled populations.
## Replaces the previous `seroincidence` package.
## Depends: R (>= 3.5.0)
## License: GPL-3
## Imports: dplyr (>= 1.1.1), magrittr, parallel, Rcpp, rlang, stats,
## tibble, tidyr, utils
## Suggests: knitr, rmarkdown, pander, Hmisc, tidyverse, fs, testthat (>=
## 3.0.0), readr, ggplot2, bookdown, doParallel
## VignetteBuilder: knitr
## LazyData: true
## Encoding: UTF-8
## URL: https://github.com/UCD-SERG/serocalculator,
## https://ucd-serg.github.io/serocalculator/
## RoxygenNote: 7.2.3.9000
## NeedsCompilation: yes
## LinkingTo: Rcpp
## Language: en-US
## Roxygen: list(markdown = TRUE)
## Config/testthat/edition: 3
## Packaged: 2023-11-21 17:05:23 UTC; runner
## Author: Peter Teunis [aut, cph] (Author of the method and original
## code.), Kristina Lai [aut], Kristen Aiemjoy [aut], Douglas Ezra
## Morrison [aut, cre]
## Maintainer: Douglas Ezra Morrison <demorrison@ucdavis.edu>
## Built: R 4.3.2; x86_64-pc-linux-gnu; 2023-11-21 17:05:27 UTC; unix
## RemotePkgRef: local::.
## RemoteType: local
##
## -- File: /home/runner/work/_temp/Library/serocalculator/Meta/package.rds
# Show citation.
citation("serocalculator")## To cite package 'serocalculator' in publications use:
##
## Teunis P, Lai K, Aiemjoy K, Morrison D (2022). _serocalculator:
## Estimating Infection Rates from Serological Data_. R package version
## 0.1.0.9000, https://ucd-serg.github.io/serocalculator/,
## <https://github.com/UCD-SERG/serocalculator>.
##
## A BibTeX entry for LaTeX users is
##
## @Manual{,
## title = {serocalculator: Estimating Infection Rates from Serological Data},
## author = {Peter Teunis and Kristina Lai and Kristen Aiemjoy and Douglas Ezra Morrison},
## year = {2022},
## note = {R package version 0.1.0.9000,
## https://ucd-serg.github.io/serocalculator/},
## url = {https://github.com/UCD-SERG/serocalculator},
## }